Background
Innovative procedures based on digital imaging are changing healthcare: clinical decision making becomes increasingly dependent on images, and images permeate all aspects of medicine from detection and diagnosis to interventions and treatment monitoring. The number of images consequently increases, increasing healthcare costs for their acquisition, but importantly also for image interpretation and communication. Automation as proposed in this project can help: firstly, by detecting the abnormal cases, and secondly by extracting relevant and characteristic parameters of the abnormal cases, relating them to known cases, and presenting them in an intuitive and interactive workflow.
Within this project, we have focused on three application areas: oncology, opthalmology, and pathology.
In oncology, radiological imaging is a crucial component of the disease pathway. Cancer patients undergo imaging for detection, diagnosis, and treatment monitoring. CT imaging of the thorax and abdomen is a large portion of the oncological workload of a radiology department and therefore, we focused on automation of different aspects of the interpretation of oncological scans.
Retinal diseases, such as age-related macular degeneration (AMD) or diabetic retinopathy (DR), are compromising the vision of more and more patients and causing blindness in an alarmingly large percentage of world population. Multimodal retinal imaging has become an indispensable diagnostic tool in Ophthalmology to early detect, treat and monitor these diseases and automated retina image analysis has the potential to improve the diagnostic process and make treatment monitoring more effective. However, efficiently leveraging information contained in multimodal imaging is a complex task and many traditional automated solutions do not suffice to accurately extract the required information. Specially, the manual assessment of quantitative temporal change for treatment decision of AMD suffers from observer dependence, and the joint assessment of different imaging techniques for one patient leave ambiguities in terms of spatial correspondence.
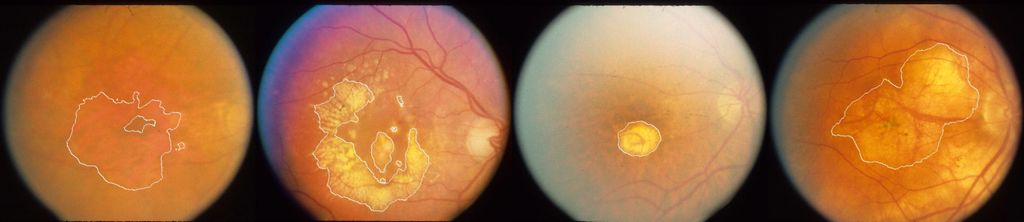
Automatic segmentation of geographic atrophy in color fundus images.
For the pathology application, the focus is on breast cancer. Breast cancer is the most common cancer among women. Within their lifetime, 12% of women are diagnosed with breast cancer. The prognosis of breast cancer patients is mainly determined by whether the cancer is organ-confined or has spread to other parts of the body. An internationally accepted means to classify the extent of cancer is the TNM staging system. In breast cancer, TNM staging takes into account the size of the tumor (T-stage), whether the cancer has spread to the lymph nodes (N-stage), and whether the tumor has metastasized to other parts of the body (M-stage). The axillary lymph nodes are typically the first location breast cancer metastasizes to. Currently, the status of these lymph nodes is almost always assessed by applying the sentinel lymph node procedure. This procedure identifies the nearest lymph nodes to which the tumor drains, which are then excised for pathologic examination. Glass slides are prepared from the tissue speciments and stained with hematoxylin and eosin (H&E) to highlight the cell nuclei and the general structural features of the tissue. Through microscopic assessment the pathologist screens the slides for tumor presence. If tumor cells are found, the pathologist measures their extent in order to determine the pathologic N stage (pNstage) of the tumor. In case of unclear diagnosis on H&E, immunohistochemical (IHC) staining for cytokeratin can be used for clarification and is standard diagnostic practice in the Netherlands.
Results
An oncology application was developed that focuses on automating the analysis of thorax-abdomen CT scans acquired from oncological patients: CIRRUS Oncology. A large database with data from more than 6000 patients has been collected at Radboudumc that helped to develop and validate the different components of the application. Within the application, novel algorithms were developed for deep-learning based thorax abdomen registration, liver, spleen and kidney segmentation and analysis, more general autosegmentation tools for other organs, and detection and quantification of bone metastases and enlarged lymph nodes. CIRRUS Oncology also allows to automatically compute RECIST reports.
An opthalmology application was developed for the automated analysis of multimodal retinal imaging examinations in order to assess treatment response in patients with advanced wet AMD and provide treatment decision support to clinicians: CIRRUS Opthalmology. Deep learning algorithms have been developed that can accurately analyze and quantify changes in follow-up optical coherence tomography scans and multimodal retinal images.
The pathology application that was developed focused on detecting and outlining the tumor areas on the digitized versions of the glass slides. These outlines help to steer the attention of the pathologist during assessment for faster, and more accurate evaluation. CIRRUS Pathology includes deep learning algoritms for automatic tissue segmentation, metastases detection, and includes a histology registration algorithm, and tools for efficient lymph node reporting.
Besides the three clinical demonstrator applications encompassing several scientific results, significant backend developments comprise challengr, a framework for algorithm validation, the DIAG grand-challenge.org, the online challenge web platform, RedLeaf, the MEVIS “Remote Deep Learning Framework”, generalized methods for image co-registration, and CIRRUS, a modular framework for locally installed and web-based clinical applications integrated with clinical backend systems, like the PACS.
Funding
AMI was a collaborative project of the Fraunhofer Gesellschaft and the Radboud University and University Medical Center and funded through the Fraunhofer ICON program.
Media
- Best Demo Award from Live Demonstrations Workshop at SPIE Medical Imaging 2017 : “A Multimodal Workstation for Analysis of Retinal Images”










