Start date: 16-09-2019
End date: 15-03-2020
Clinical problem
Immunotherapy with immune checkpoint inhibitors (ICI) has led to unprecedented responses in previously untreatable cancers, including non-small cell lung cancer (NSCLC). Unfortunately, only one out of five patients responds to immunotherapy, and the other patients are exposed to drug toxicity without therapeutic effect, at a yearly cost of more than hundred thousand euros. Current clinical guidelines select patients who will receive the treatment based on visual histological assessment of cells positive to PD-L1 via immunohistochemistry. Lung pathologists have to visually estimate the proportion of PD-L1 positive tumor cells versus all tumor cells, in tissue samples containing hundreds of thousands of tumor cells. This subjective and poorly reproducible procedure comes with a high inter-observer variability in assessment of PD-L1 positive cells, and both clinical trials and clinical practice have shown that using the current procedure, not only most selected patients do not respond (around 35-40% of selected patients respond), but some of the untreated patients (around 15%) would have responded to the immunotherapy if they had been treated.
Innovation
The goal is to develop computational pathology models based on artificial intelligence, to assist detect tumor and immune cells in lung cancer specimens and classify them into PD-L1 positive and negative. The aim is to create the basis for a tool that can be used in the clinic by pathologists to assess biomarkers based on PD-L1 staining on gigapixel histopathology whole-slide images in an objective and reproducible way.
Results
Three different neural network models were tested for cell detection, namely VGG-like network with 11 layers, DenseNet, and U-net. In all cases, models were trained with data from a cohort of of 29 whole slide images of resected lung cancer specimens (50% adenocarcinoma, 50% squamous cell carcinoma). In each image, an expert annotated positive and negative cells in selected regions of interest using point annotations. For NatureNet and DenseNet, training was done with patches that have been cropped around annotated cells from the training set. For the U-net model, segmentation masks were created around the original point annotations and used to train the segmentation model end-to-end. A novel sampling strategy that only considers pixels around the annotations was developed.
Cell detection and classification performance were assessed using the F1-score for five classes, namely 1) PD-L1 positive tumor cells, 2) PD-L1 negative tumor cells, 3) PD-L1 positive immune cells, 4) PD-L1 negative immune cells, and 5) other cells plus background tissue (which in practice corresponds to the “rest” of tissue components in the image). The VGG-like network showed the best F1-score for the cell classification compared to the other models explored in this project: VGG-like F1=0.676, DenseNet F1=0.582, U-net F1=0.559. The overall results on the test set in the form of a confusion matrix are summarized in the figure below. Overall, we observed that the network shows fairly good performance at differentiating between PD-Ll positive and negative cells but have problems differentiating between positive tumor and immune cells or negative tumor and immune cells.
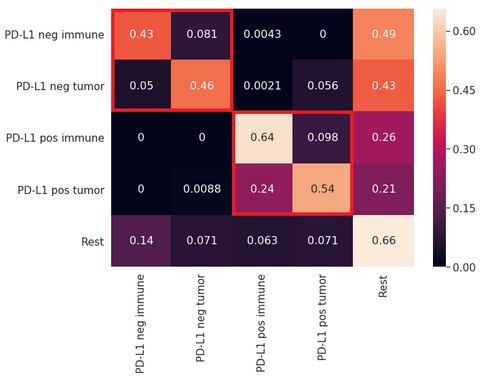
Confusion matrix showing the performance of the U-Net model developed in this project.
At the same time, we found that the U-net model produced the best visual segmentation performance of all the developed networks, which makes it easier for pathologist to interpret the results. For this reason, we selected U-Net as the model to be implemented in the form of a web-based application, which can be found here. The figure below depicts an example of a lung cancer slide stained with PD-L1 and processed with the U-Net model developed in this project. It can be noted that regions with brown staining, containing PD-L1 positive cells, are mostly detected as such (in green color immune cells and in yellow tumor cells), although we observe the presence of both false positives and false negatives. PD-L1 negative cells are also found outside the brown region, but false positives and false negatives are observed here as well. In all cases, cell nuclei are identified but not precisely segmented, mostly because of the way the segmentation masks were created from point annotations to train the U-Net model.
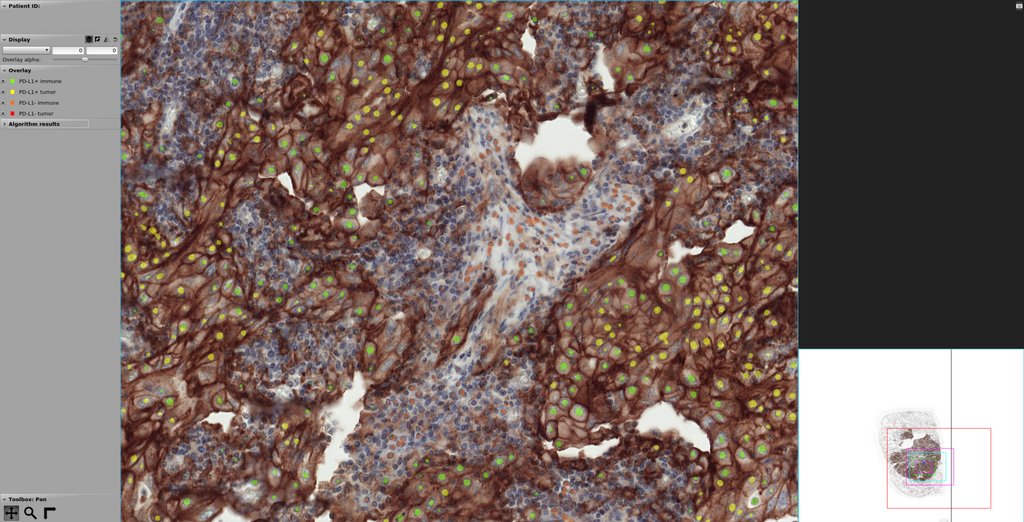
Screenshot of the web application for segmentation of PD-L1 positive and negative cells.
In conclusion, in this project, we have developed a first prototype to address the problem of automated PD-L1 quantification of positive and negative tumor and immune cells in lung cancer histopathology images. While the results are not ready for clinical application of the algorithm, this work represents a starting point for future work and a baseline approach to use in comparison with other methods.
The web application developed in this project based on grand-challenge can be found here:
The code for this project can be found in this GitHub repository. The final report for this project can be found here.


