Start date: 01-05-2021
End date: 07-04-2022
Clinical problem
Recent studies have developed our knowledge regarding the tumor microenvironment to exploit tumor immunity in fighting breast cancer (BC). An important part of the immune system response to invase tumors is formed by tumor infiltrating lymphocytes (TILs), which are immune cells that infiltrate tumor tissue. Multiple studies have established a positive relationship between higher levels of TILs and survival outcomes in Triple Negative Breast Cancer (TNBC) and human epidermal growth factor receptor 2 positive (HER2+). To estimated the amount of TILs, the International TILs working group has established recommendations based on the following steps: 1. Define the tumor area in which TILs are to be evaluated 2. Identify the region of the slide containing tumor-associated stroma (i.e., connective tissue) 3. Only evaluate mono-nuclear TILs 4. Asses the percentage of TILs as the fraction of stromal tissue covered by them
Scoring the percentage of TILs in a slide typically requires a trained pathologist to perform the evaluation. As such, manual TIL evaluation from slides is a slow process that can only be performed by a limited set of people. Automatically calculating the TIL score as well as producing detection and segmentation maps of stromal areas and their respective lymphocytes could be of great benefit to TIL research as well as facilitate the implementation of TIL scoring in clinical practice. This project aims to develop a system which segments the stromal area, detects and segments the TILs present within this area and subsequently uses this map to calculate a TIL score per slide.
Solution
The purpose of this project is to produce an automated pipeline that can reproduce all of the steps involved in TIL scoring. First, the tumor including the invasive edge will be segmented, within this segmentation the stromal area will be marked as the proposed region in which to locate and count the TILs. After this step the individual TILs will be detected and segmented. Lastly we will automatically generate a TIL score per slide. The automatic calculation of the TIL score should considerably reduce pathologist efforts in diagnostics while the segmentation map is a tangible output that provides interpretability and allows for convenient checking of the TIL score.
Data
The main source of data for this project was the provided by the TIGER dataset which was released as part of the challenge by the same name (link below). The challenge provided annotations for both tissue segmentation and lymphocyte detection as well as the TIL-scores estimated by a pathologist. Additionally, trough the Grand Challenge platform the challenge allowed the evaluation of predicted TIL scores on their predictive value of patient survival time. During model development the NuCLS dataset was used as an additional source of data while waiting for the full TIGER dataset to be released.
Approach
Cyril will be supervised by Francesco, Mart, and Witali whom all have previously worked on research related to TIL detection, segmentation and scoring. To create a system that can perform stroma segmentation, TIL detection and automated TIL scoring the state of the art convolutional neural approaches will be reviewed and adapted to the problem at hand. The student gets access to the high-performance deep learning cluster of DIAG, named SOL, for running machine learning experiments.
Methods
We propose a multi-part system as a solution to this problem that uses AI to tackle of the steps required in TIL-scoring. A Hooknet segmentation network was trained the separate the tissue types into invasive tumor, tumor-associated stroma and rest. Methods from computational geometry were combined with kernels to determine the bulk of the tumor. A Faster R-CNN model was trained for the task of TIL detection. We subsequently detect TILs only within the tumor-associated stroma that falls within the border of the tumor bulk. A Faster R-CNN model was trained for the task of TIL detection. Lastly, different methods of quantifying the detected TILs were experimented with. These included, taking the ratio of the area of the TILs and the tumor bulk area, taking the same ratio but only within the margin of the tumor, using the ouputs of the segmentation model to determine how much of the tumor-associated stroma was infiltrated by the lymphocytes.
The full TIL-scoring procedure is visualised in the image below:
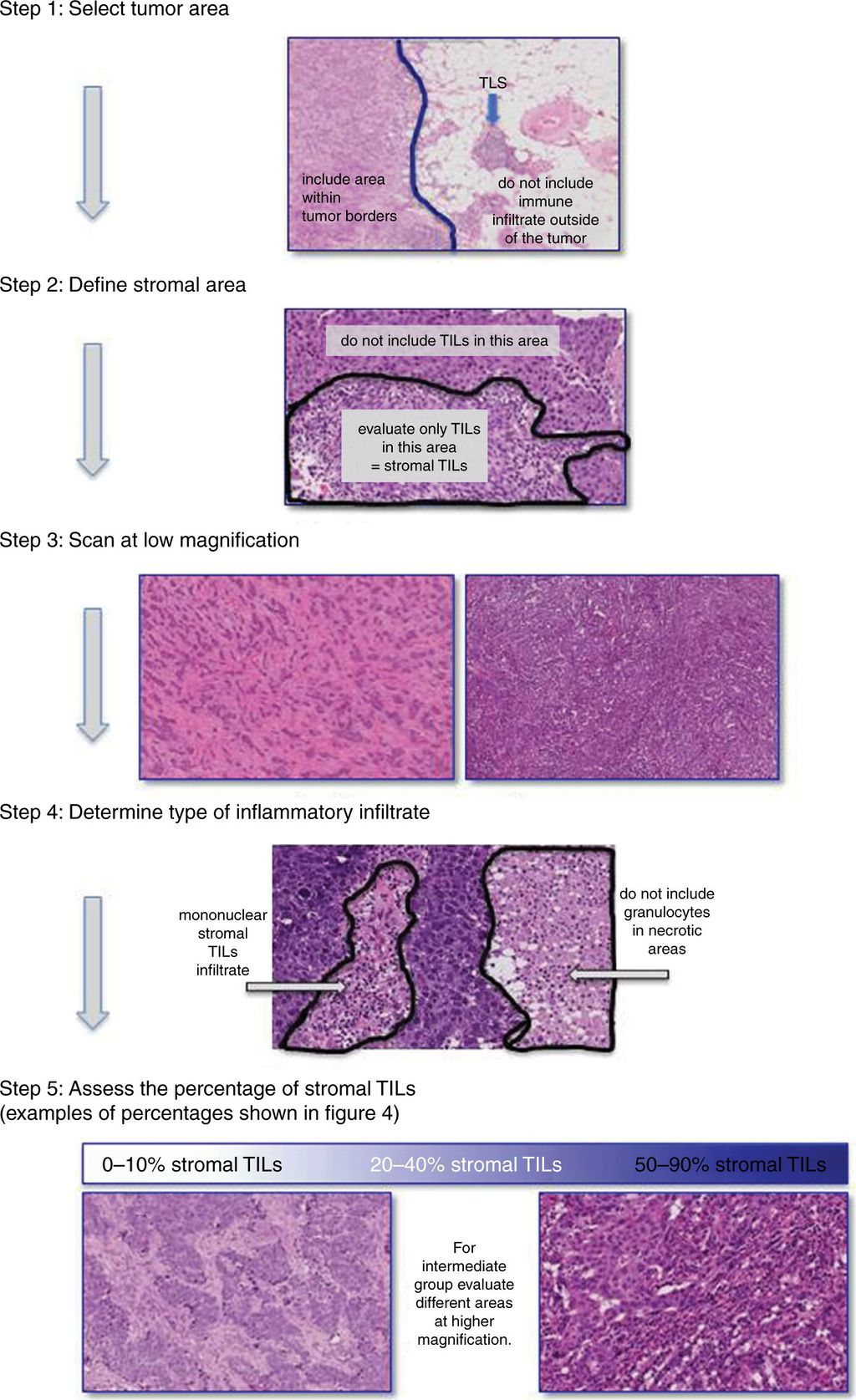
Results
The Segmentation network achieved a overall Dice score of 0.72 on a test set. The TIL detection model achieved a FROC score of 0.179. The TIL quantification methods were evaluated by their correlation with the scores assigned by a pathologist and their predictive value for patient outcome. Calculating a TIL-score using only the lymphocytes found within the margin of the tumor bulk resulted in the highest predictive value with a C-index of 0.704, which was an improvement over the C-index of 0.63 using clinical variables. The method had Spearmans correlation of 0.56 with the estimates of a pathologist.
Conclusion
In conclusion, it can be seen the recommendations and guidelines of the International TILs working group can be implemented in an automated framework for TIL-scoring. The developed approach shows moderate correlation with pathologist estimates as well as an improvement in prognostic value over the medical baseline. Different TIL-scoring methods varied little in their correlation with pathologists or medical outcomes. Lastly, the findings of this thesis suggest automated TIL scoring methods could assist pathologists in their daily work.
The algorithm was used as the baseline for the TIGER challenge which can be found on the Grand Challenge platform.
The source code to the algorithm can be found on Github.




