The Gleason score is the most important prognostic marker for prostate cancer patients but suffers from significant inter-observer variability. We developed a fully automated system using deep learning that can grade prostate biopsies following the Gleason Grading System. A description of our method and research can be found in the paper: Automated deep learning system for Gleason grading of prostate biopsies: a development and validation study. A summary is available in a blog post on our study.
The algorithm we developed is available to try out online, without any requirements on deep learning hardware.
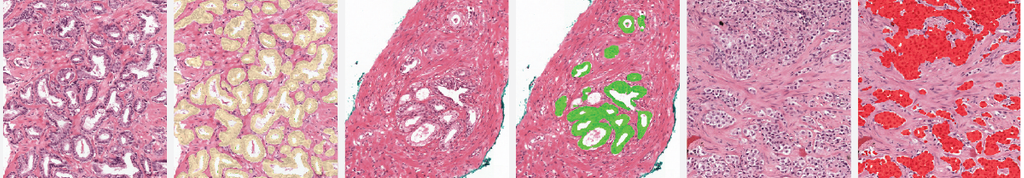
Online examples
The two examples below show the raw output of the algorithm, without any post-processing, overlayed on biopsies. Benign tissue is colored green, Gleason 3 in yellow, Gleason 4 in orange and Gleason 5 in red. The original biopsy is shown on the left, the biopsy with overlay on the right. You can zoom in by scrolling, moving around can be done with click&drag.
Trying out the algorithm
Our trained system can be tried out online through the Grand Challenge Algorithms platform. All processing is performed on the platform; there is no specialized hardware required to try out our algorithm. Running the algorithm requires an account for Grand Challenge. If you don't have an account yet, you can register at the website; alternatively, you can log in using a Google account. After registering for a new account, or logging in to an existing Grand Challenge account, you can request access to the algorithm.
The algorithm can be run with a minimum of one biopsy, supplied in a multiresolution tiff file. For more details on how to generate such a file, see below. We assume that the following magnification levels are available in the slide (measured in μm pixel spacing): 0.5, 1.0, and 2.0 (with a small tolerance). Note that the algorithm will provide two outputs, the first is the normalized version of the uploaded image, the second is the output of the Gleason grading system. To visualize the output of the Gleason grading system, press the second 'View input & output' button. You can download the processed results as well.
Before submitting, please take our Terms & Conditions in to account.
Description of the pipeline
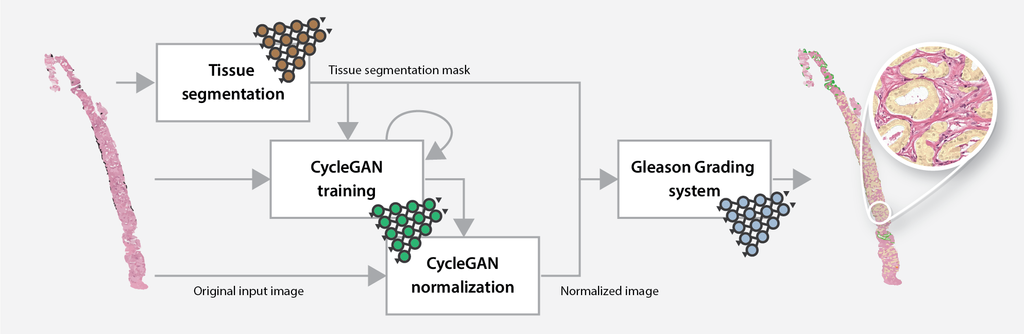
The online Gleason grading algorithm consists of multiple steps to prepare and analyze the data:
- For each biopsy, a tissue mask will be generated by a deep learning-based tissue segmentation system.
- Based on the input and a small set of target files, a CycleGAN-based normalization algorithm is trained.
- The trained normalization algorithm is applied to all input biopsies to normalize the data.
- Each normalized biopsy is processed by the automated Gleason grading system. The output of the system consists of an overlay and a set of metrics for each biopsy. The metrics include the biopsy-level grade group, the Gleason score, and predicted volume percentages.
Each job roughly completes within an hour, depending on the size and number of biopsies, and the processing capacity. Jobs are processed on a first-come, first-served basis. The actual Gleason grading system itself can grade a biopsy within a few minutes, the remainder of the time is spent on the normalization task. For real world scenarios, this preprocessing step is not needed or can be pre-trained to speed up the process.
The training time of the normalization algorithm is independent of the number of biopsies. It is, therefore, more efficient to submit several biopsies at once, than to submit multiple jobs with a single biopsy.
Terms & Conditions
When submitting data to the algorithm, please take the following into account:
- The use of the algorithm is meant for research purposes only. The algorithm was trained on data from a single center, and can give the wrong diagnosis on external data. We give no assurance on the correctness of the system's output.
- A custom normalization algorithm is trained on the input data, though this cannot overcome all stain and scanner differences. We limit the overall processing time of each job by setting an upper bound on the number of epochs the normalization algorithm is trained. Due to the limited processing time, in some cases the normalization technique can fail.
- We currently only support multiresolution .tif files as input.
- All input images should contain magnification levels that correspond to a 0.5, 1.0, and 2.0μm pixel spacing (± 0.05). The algorithm will stop if any of the input images miss one or more of these levels.
- Following the Terms & Conditions of the Grand-Challenge platform, submitted data can be used for future research projects. We assume that submitted data has been anonymized and that you, as a submitter, have the right to submit the image.
More information
Please refer to any of the following sources for more information:
- A summary of the research can be found in our blog post.
- The paper can be read in the Lancet Oncology, a preprint is available on arXiv.
- This algorithm is part of the Deep PCA research project.
- To generate a multi-resolution TIF file from proprietary WSI scanner formats, we recommend using the multiresolutionimageconverter command line tool which is provided with the ASAP package.
Further questions regarding the Gleason grading system can be addressed to Wouter Bulten.








