Problem
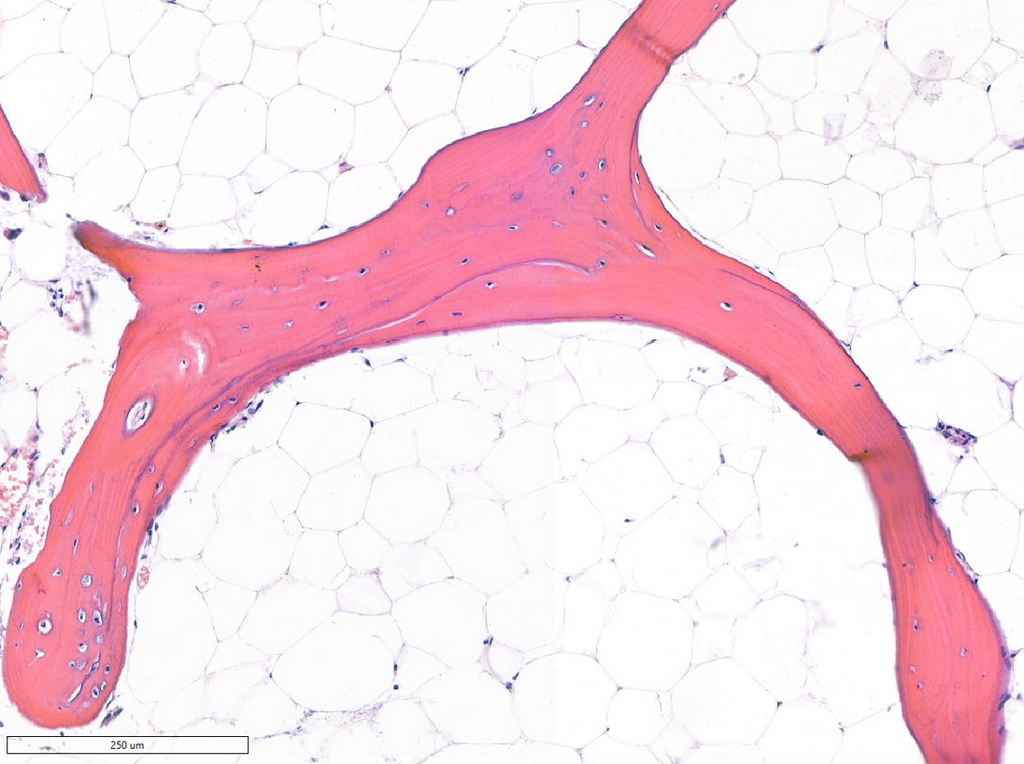
Bone is a highly organized tissue for which the spatial arrangement of cells relative to the mineralized matrix determines its mechanical and healing properties. Understanding how this organization varies between individuals or under different biological conditions could reveal key principles of bone adaptation and regeneration.
Despite the availability of high-resolution digital slides, there is currently no standardized computational approach to quantify cell-to-cell and cell-to-matrix spatial organization in bone tissue. Bridging this gap would enable data-driven links between tissue architecture and patient characteristics, as an emerging frontier in computational pathology.
This project aims to develop an AI-driven spatial analysis toolkit that quantifies how cells are organized within bone tissue and how this structure changes across patient groups.
Approach
You will develop or adapt algorithms to describe the spatial topology of bone microarchitecture from digital pathology data. Building on existing nuclei point annotations and bone segmentation masks, you will compute features capturing:
- Cell-to-cell organization, such as clustering, nearest-neighbor distances, and spatial autocorrelation.
- Cell-to-matrix relationships, including osteoblast alignment along bone surfaces.
- Aggregate spatial metrics summarizing tissue organization at the slide and patient level.
You will then apply statistical and machine learning methods to explore correlations between these spatial features and biological variables (e.g., age, sex) to advance a framework for interpretable AI-based histomorphometry.
Data
The dataset consists of more than 300 anonymized, H&E-stained, human bone images with accompanying bone segmentation masks and point-level nuclei annotations. This curated dataset enables large-scale spatial feature extraction and comparative analysis across patient groups.
Requirements
- Master’s student in Artificial Intelligence, Computer Science, Biomedical Engineering, or a related field.
- Proficiency in Python and experience with scientific computing (NumPy, SciPy, scikit-learn, Pandas).
- Familiarity with image analysis, spatial statistics, or graph-based feature extraction is highly desirable.
- Interest in applying algorithmic methods to biomedical imaging; no wet-lab work required.
Information
- Project duration: 6–9 months
- Location: Radboud University Medical Center, Nijmegen, The Netherlands
- You will join the Computational Pathology Group, a leading research team for deep learning in digital pathology. The project is supervised by Gerry Koons, Marie Skłodowska-Curie Postdoctoral Fellow, and will provide access to a large GPU cluster, high-resolution pathology data, and interdisciplinary mentorship bridging AI, pathology, and regenerative medicine.
- For more information, please contact Gerry Koons.

