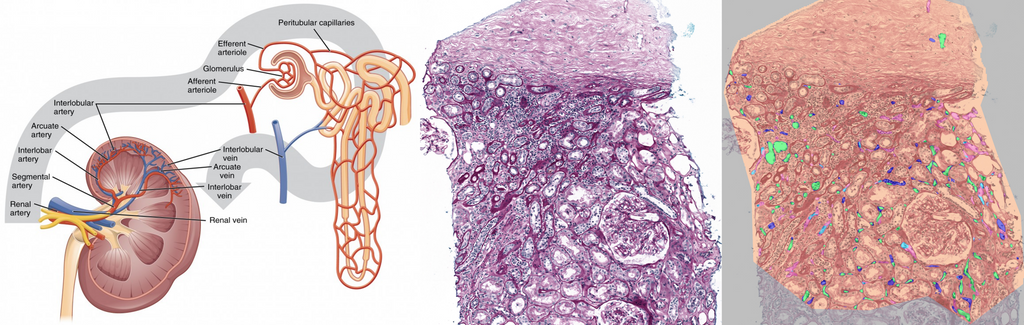
Clinical Problem
Peritubular capillaritis (ptc) refers to inflammation of the peritubular capillaries (PTCs), which are the tiny blood vessels surrounding the renal tubules (which contain the urine) in the kidneys. They ensure efficient reabsorption and secretion of substances between the bloodstream and tubules. Inflammation of peritubular capillaires is a sign of microvascular damage. Assessing the presence and degree of PTCs is an important feature for diagnosing antibody-mediated rejection (ABMR) in kidney transplants. This task is tedious for pathologists, as they must search for individual inflammatory cells within PTCs, decide if more than 10% of the surface is affected, and count the number of inflammatory cells within the most affected peritubular capillary. It suffers from poor to moderate interobserver variability. To automate this process, we have developed a first algorithm for PTCs segmentation. However, certain tissue regions remain challenging (e.g. high inflammation and atrophic tubules), and thus leave room for improvement.
Approach
Our current approach is based on a U-Net segmentation model using a ResNet50 backbone. In this project we will investigate other state-of-the-art approaches, specifically instance segmentation, to improve performance. Examples of this are SAM[3] and DINO[4]. We currently also do not use any post-processing, so this is another area to explore. Additionally, there is also the opportunity for a clinical analysis of the PTCs characteristics in relation to patient data (shape, size, etc.).
Data
So far, we have collected a dataset of 69 Whole Slide Images (WSI) of kidney transplant biopsies. On these slides, a resident pathologist annotated individual PTC in several regions of interest per slide. Regions of interest are present for both the medulla and cortex region of the kidney. More un-annotated slides are available to use, as well as the possibility to work with a pathologist to acquire more annotations, especially of difficult regions. The patient data of this cohort is also well-characterized and available.
References
[1] Jacq, Amélie, et al. "Automated evaluation with deep learning of total interstitial inflammation and peritubular capillaritis on kidney biopsies." Nephrology Dialysis Transplantation 38.12 (2023): 2786-2798.
[2] Chen, Yijiang, et al. "Clinical relevance of computationally derived attributes of peritubular capillaries from kidney biopsies." Kidney360 4.5 (2023): 648-658.
[3] Zhang, Jingwei, et al. "SAM-path: A segment anything model for semantic segmentation in digital pathology." International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer Nature Switzerland, 2023.
[4] Liu, Shilong, et al. "Grounding dino: Marrying dino with grounded pre-training for open-set object detection." arXiv preprint arXiv:2303.05499 (2023).
Requirements
- Students with a major in computer science, biomedical engineering, artificial intelligence, physics, or a related area in the final stage of master level studies are invited to apply.
- Interest in image analysis and machine learning.
- Affinity with programming in Python, specifically in the PyTorch framework
Information
- Project duration: 6 months
- Location: Radboud University Medical Center
- You will be part of the Diagnostic Image Analysis Group (DIAG) and Computational Pathology Group, whose research focus is the analysis of histopathological slides with deep learning techniques.
- You will have access to and work with a large GPU cluster.
- For more information, please contact Linda Studer


