Clinical Problem
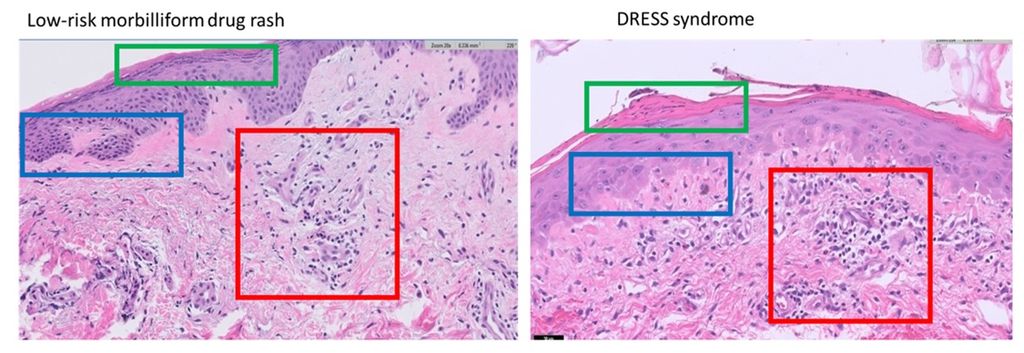
Drug reaction with eosinophilia and systemic symptoms (DRESS) syndrome is a syndrome resulting from an adverse drug reaction. It can cause liver damage, kidney damage, as well as many other systemic symptoms including a rash. Low-risk morbilliform drug eruptions are another kind of adverse drug eruption, and while the rash it presents with is similar in appearance to the rash of DRESS, it does not come with the dangerous systemic symptoms. Rendering the proper diagnosis in a timely fashion is imperative as DRESS syndrome can carry a mortality rate of up to 10% (Hama et al, 2022). Both DRESS syndrome (Choudary et al, 2013) and low-risk morbilliform drug eruptions (Walman et all, 2022) present with non-specific morbilliform rashes. Further, hospitalized patients are often acutely ill, which makes differentiating these two diagnoses based on objective criteria like fevers and organ involvement difficult. For the same reasons, laboratory tests have also been shown to be of limited utility (Ernst et al, 2022). Given the limitations in using clinical and laboratory parameters to differentiate these diagnoses, histopathologic examination has become an attractive avenue. However, histopathologically, morbilliform drug eruptions are characteristically ambiguous and non-specific (Ernst et al, 2022).
Solution
In our study, we aim to utilize skin biopsy samples of patients who received a definitive diagnosis of either DRESS syndrome or low-risk morbilliform drug eruption. We will collate the samples, and work to train an AI algorithm to help identify distinguishing factors between them. We hypothesize that the AI will be able to differentiate between DRESS syndrome and low-risk morbilliform drug eruptions, helping to provide us with an additional tool in our ability to differentiate these conditions. After we train the AI retrospectively on already-diagnosed samples, we plan to employ it prospectively to see if it can readily distinguish the two eruptions.
Approach
The student will be supervised by a research member of the Diagnostic Image Analysis Group and Computational Pathology Group whose research is dedicated to analyses of histopathological slides with deep learning techniques. The student will have access to a large GPU cluster.
Requirements
- Students with a major in computer science, biomedical engineering, artificial intelligence, physics, or a related area in the final stage of master level studies are invited to apply;
- affinity with programming in Python;
- interest in deep learning and medical image analysis.
Information
- Project duration: 6 - 9 months
- Location: Radboud University Medical Center
- For more information, please contact Geert Litjens
