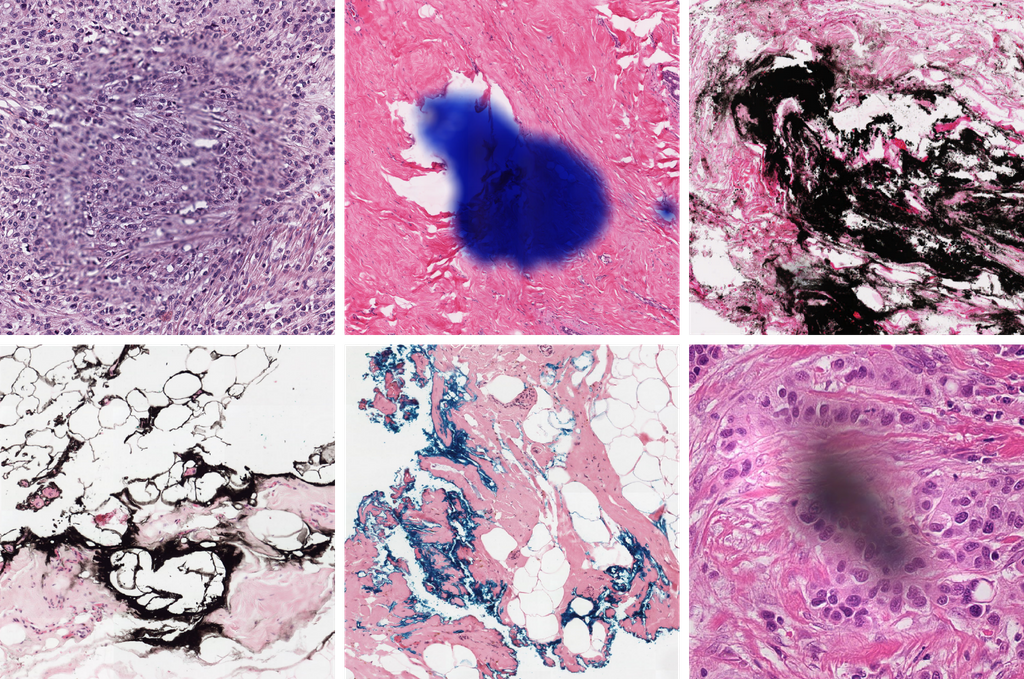
Background
Whole slide imaging technology allowed the digitization of conventional glass slides which led to several new opportunities in the field such as the integration of computational systems, most notably artificial intelligence. However, the digitization process also brought along new challenges for the automated analysis of tissue images. An important challenge arises from the presence of image artifacts, including, but not limited to - out of focus regions, - regions containing ink, - tiling effect due to the stitching, - folded or torn regions, or completely missed regions during scanning. Accurate analysis of whole slide images depends on detection and elimination/correction of such artifacts.
Research question: Is it possible to reliably detect the artifacts in whole slide images?
Tasks
The main goal of this project is to develop a deep learning model that can detect and classify the different types of artifacts observed in whole slide images.
Requirements
- Students with a major in computer science, biomedical engineering, artificial intelligence, physics, or a related area in the final stage of master level studies are invited to apply.
- Interest in image analysis and machine learning.
- Affinity with programming in Python is required.
Information
- Project duration: 6 months
- Location: Radboud University Nijmegen Medical Center
- For more information please contact Caner Mercan.
