Clinical Problem
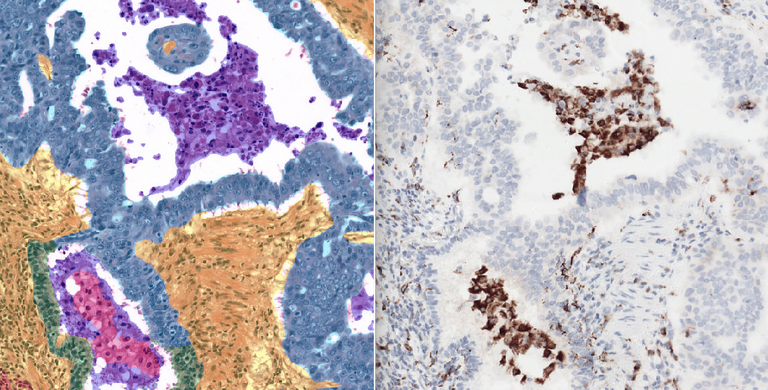
Non-Small Cell Lung Cancer (NSCLC) continues to pose a significant challenge in cancer treatment, despite the considerable promise of immunotherapy, which can be highly effective. Unfortunately, current biomarkers used to assess whether a NSCLC patient will repond to immunotherapy are far from optimal. To improve the stratification of responders and nonresponders for immunotherapy, we need a better understanding of the interaction between the patients' immune system and the tumor, known as the Tumor-Immune Micro Environment (TIME). The realm of digital pathology provides a valuable platform for delving into the TIME, enabling us to identify patterns in both the tumor and immune cells and how they interact. Increasingly, scientific literature highlights the vital role played by macrophages in the Tumor-Immune Micro Environment (TIME), and their relation to immunotherapy response. In research settings, CD68 immunohistochemically stained slides are typically employed to evaluate the presence of macrophages, but this marker is not routinely assessed in clinical practice. However, macrophages are also visible in clinical standard H&E stained slides.
Solution
The Computational Pathology Group (CPG) of Radboudumc has developed an artificial intelligence algorithm to segment various tissue classes in NSCLC tissue, including tumor tissue and macrophages in H&E-stained histopathology. While this model already excels in overall tissue segmentation performance, it performs suboptimally at identifying macrophages because of subjective training data used for development and its limited size. Therefore, the potential utility of macrophages as a biomarker and their predictive or prognostic value have yet to be explored. In this project, we will leverage the state-of-the-art nnUNet segmentation framework, recently tailored to pathology by our group, and use it to improve the model's ability to identify macrophages accurately. Additionally, we aim to convert the model's output into clinically explainable biomarkers, such as macrophage densities and their localization in the TIME.
Approach
The project provides comprehensive support, including pre-existing code, commented examples for nnUNet utilization, and a framework for creating new training data from existing segmentation masks. The nnUNet model also generates inference uncertainty maps, offering potential areas for further improvement beyond IHC guidance. The candidate will be encouraged to investigate the correlation between macrophages and clinical questions and transform segmentation model output into insightful biomarkers.
Data
Our project benefits from a rich dataset comprising 60 H&E slides and corresponding (serial section) slides stained with CD68 Immunohistochemistry (IHC), a reliable macrophage marker. This dataset allows precise identification of macrophage locations in the H&E slides therefore generating a large-scale objective reference standard for model development and validation. Additionally, we offer access to multiple bigger datasets for exploring and assessing macrophage biomarkers for clinically relevant endpoints.
Requirements
- Students with a major in computer science, biomedical engineering, artificial intelligence, physics, or a related area in the final stage of master level studies are invited to apply.
- Affinity with programming in Python, specifically in the PyTorch framework.
- Interest in deep learning and medical image analysis.
Information
- Project duration: 6 months
- Location: Radboud University Medical Center
- For more information, please contact Joey Spronck

