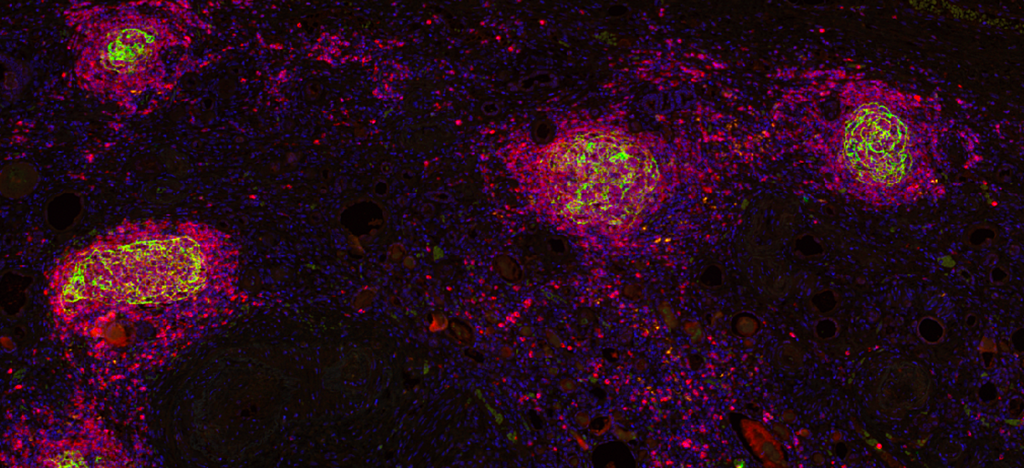
Clinical Problem
Every day, oncologists must decide how cancer patients should be treated. The increasing amount of available treatment options, ranging from chemotherapy, radiotherapy, immunotherapy, targeted therapies, and surgery, does not make this choice an easy one, and there is no one-size-fits-all solution because every patient is different.
In recent years, an increasing number of biomarkers have been proposed, aiming at predicting the chances that patients will respond to different types of treatment and to predict prognosis.
In non-small cell lung cancer (NSCLC), the deadliest cancer, the role of immune cells and their interaction with tumor cells have been investigated and encoded into biomarkers that could be used in the clinic. Among them, the presence of tumor-infiltrating lymphocytes (i.e., immune cells that can potentially attach and destroy tumor cells) and of tertiary lymphoid structures (i.e., organized aggregates of immune cells that appear in the proximity of the tumor) have been shown to correlate with treatment response and patient survival. Although these biomarkers could potentially encode complementary mechanisms of the tumor-immune interaction, so far, they have been mostly considered as independent biomarkers.
The Computational Pathology Group (CPG) of Radboudumc has developed artificial intelligence algorithms to quantify TILs and TLS in H&E-stained histopathology slides of multiple tissue and cancer types and shown the predictive and prognostic value of these biomarkers in NSCLC. But the potential of an AI-based approach to simultaneously assess TILs and TLS (T&T) and the analysis of their joint predictive and/or prognostic value has not been explored yet.
Solution
In this project, we will start with the existing AI algorithms for TILs and TLS developed within the CPG and investigate ways to combine them both by learning them at the level of whole-slide image analysis via weakly-supervised learning, and by combining them into statistical predictive models. We aim at assessing the predictive and prognostic value of a novel T&T biomarker and at building an AI model to efficiently extract it from histopathology whole-slide images of NSCLC.
Data
At Radboudumc, you will be working on the GENERATOR dataset of early-stage NSCLC, consisting of 500 cases with survival data. This is a multimodal dataset consisting of digital pathology slides of pre-operative biopsies and resected tumor sections, plus CT and PET scans of the tumor, allowing to investigate the role of T&T in both biopsies and resections, and their correlation, also with CT- and PET-based features. The IGNITE dataset of late-stage NSCLC consisting of about 400 tumor biopsies of patients treated with immune checkpoint inhibitors, allowing to assess the predictive power of T&T in the context of immunotherapy treatment.
From public sources, you will have access to NSCLC data from the PLCO trial (450 patients), the CPTAC dataset (400 patients), the NLST trial (400 patients), and TCGA data (2000 patients).
Approach
The student will be supervised by a research member of the Diagnostic Image Analysis Group and Computational Pathology Group whose research is dedicated to analyses of histopathological slides with deep learning techniques. The student will have access to a large GPU cluster.
Requirements
- Students with a major in computer science, biomedical engineering, artificial intelligence, physics, or a related area in the final stage of master level studies are invited to apply;
- Affinity with programming in Python, specifically in the PyTorch framework; affinity with statistical analysis packages such as R is a plus.
- Interest in deep learning and medical image analysis.
Information
- Project duration: 6 months
- Location: Radboud University Medical Center
- For more information, please contact Francesco Ciompi
