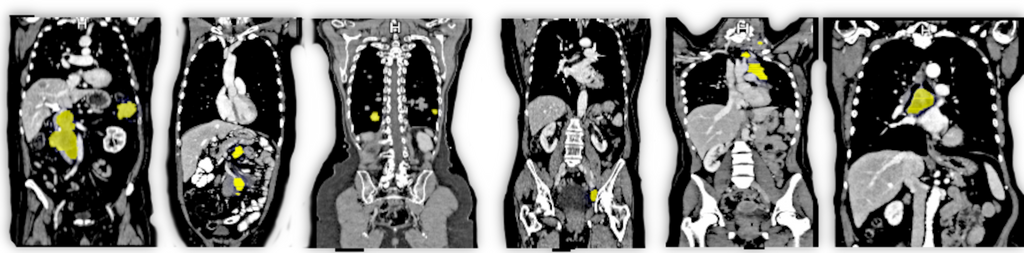
Clinical Problem
Computed Tomography (CT) scans provide a comprehensive 3D view of the body's interior and serve as a primary imaging modality for detecting metastasis – malignant growths that often hint at the spread of cancer. Given their minute size, often just a few millimeters across, metastases present a considerable challenge to discern amidst the multitude of CT image slices. Currently, expert radiologists dedicate significant time and effort to interpret these scans. The aim of automatic metastasis detection, therefore, emerges not just as a quest for accuracy but also an avenue to optimize valuable human resources in the medical domain.
Solution
To develop an AI-based model for automatic and efficient detection of metastasis in CT images. While automatic lesion detection models have been studied before, only few studies have explored the potential of utilizing expansive datasets with weak annotations. This project aims to use the vast amounts of weak annotated datasets (e.g. DeepLesion dataset) and diagnostic reports to improve the efficiency and accuracy of metastasis detection algorithms.
By reducing the need for exhaustive manual annotations and expert intervention, this approach could pave the way for a new era in medical imaging where clinicians are aided by intelligent systems, making diagnostic procedures faster and more precise.
Approach
Primary goal: Image-based Metastasis Detection - Development: Build a baseline detection model to detect metastasis in CT images, using a mixture of fully and partially annotated training data. - Evaluation: Evaluate the model using metrics like accuracy, sensitivity, specificity, and the area under the ROC curve.
Secondary goal: Incorporating Radiology Reports - Self-supervised Learning: Use information extracted from radiology reports (such as lesion type, location, size) to guide the training of the image-based model, providing it with context and helping it focus on regions of interest. - Extension & Integration: Extend the baseline model to incorporate insights gained from the diagnostic reports. - Evaluation: Assess the integrated model on a separate test set to ensure its generalizability and compare it to the performance of the purely image-based model.
Data
For this project we will have three dataset available, one from Radboudumc, while the other two publicly available. Those resulting into 42,128 lesions from 8,770 patients.
Requirements
- Students with a major in computer science, biomedical engineering, artificial intelligence, physics, or a related area in the final stage of master level studies are invited to apply.
- Affinity with programming in Python, specifically in the PyTorch framework.
- Interest in deep learning and medical image analysis.
Information
- Project duration: 6 months
- Location: Radboud University Medical Center
- For more information, please contact Max de Grauw

