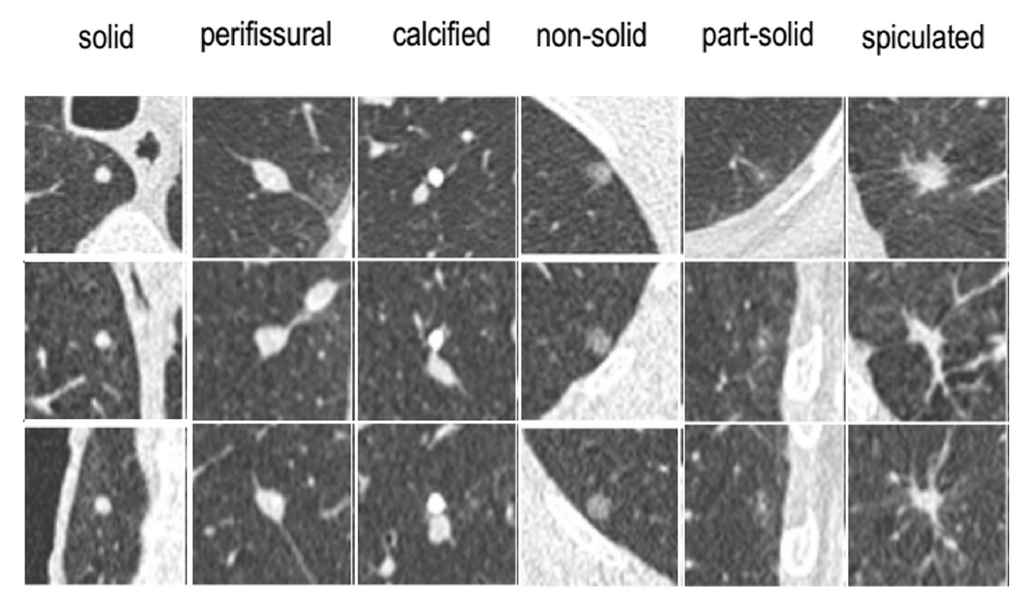
Clinical problem
Lung cancer is the leading cause of cancer death among both men and women, accounting for nearly 25% of all cancer deaths. While lung cancer typically shows up as pulmonary nodules on CT examinations, most nodules are benign and do not require further clinical workup. However, radiologist workload is expected to drastically increase soon with the widespread implementation of lung cancer screening programs. Therefore, accurate detection and characterization of pulmonary nodules are crucial for optimizing screening.
The Diagnostic Image Analysis Group (DIAG) at Radboudumc has brought Veolity to the market with MeVis Medical Solutions (Bremen, Germany). Veolity is a dedicated software solution for efficient reading of chest CT examinations in lung cancer screening programs. This product is actively used by several sites in North America, Europe, Asia, and Australia. The software includes algorithms to detect, classify, and segment pulmonary nodules.
In previous publications, we have analyzed the performance of a deep learning system for nodule classification on two screening datasets from Italy and Denmark (paper). It is at present unclear how this deep-learning based classification system performs on heterogeneous CT data collected from routine clinical practice.
Solution
You have two main tasks within this project:
- Train and improve deep learning algorithm(s) to automatically classify pulmonary nodules.
- Validate performance on an extensive set of clinical CT scans.
You will begin by taking the previously published nodule type classifier and retrain the system with more modern convolutional neural networks, preferably starting with the ResNet50 and I3D based nodule malignancy classifier described here. Then, you will perform external validation with unseen datasets once the development is frozen. Finally, you will analyze the weak points of the trained system and explore techniques how to improve performance.
Data
DIAG has a scientific archive of over 100,000 chest CT examinations including data from several lung cancer screening trials. For this project, we have curated images and labels for:
- 1,352 nodules from MILD
- 453 nodules from DLCST
- A clinical set of nodules from 100 CT scans at Radboudumc and 100 CT scans from JBZ, extensively annotated by 5 thoracic radiologists
- ~1200 nodules from LIDC/IDRI, extensively annotated by 4 radiologists
Results
The algorithm(s) that come out of this project will be made publicly available for research use as Docker containers through the grand-challenge.org platform, and may be integrated with CIRRUS Lung Screening (the research prototype version of Veolity). You are encouraged to write a scientific publication describing your results.
Embedding
You will be embedded in the Diagnostic Image Analysis Group and will be supervised by a research member whose research is dedicated to AI for lung cancer. We have a strong collaboration with both clinical and radiological experts. You will also have access to a Deep Learning GPU Cluster (SOL) to run deep learning experiments.
Requirements
- Students Artificial Intelligence, Data Science, Computer Science, Bioinformatics, Biomedical Engineering or similar in the final stages of their Master education.
- You should be proficient in Python programming and have a theoretical understanding of deep learning architectures.
- Experience with medical images is beneficial.
Information
- Project duration: 3, 6 or 9 months
- Location: Radboud University Medical Center
- For more information or to apply for this project, please contact Colin Jacobs


